Enzymes are proteins which catalyse (speed up) metabolic reactions. Like all other catalysts (e.g. in chemistry), enzymes achieve this by lowering the activation energy (energy needed for a reaction to occur) of a reaction, by forming an enzyme-substrate complex.
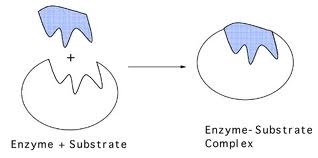
This can be described by the lock and key, and induced fit models of enzyme action. The lock and key model is based on complementary shapes between the enzyme and substrate. The substrate fits into the enzyme.
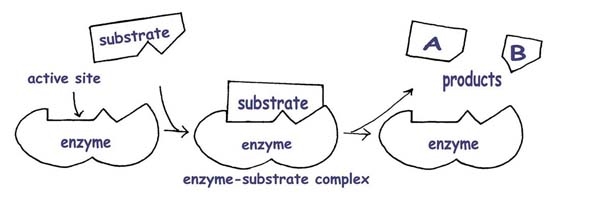
The induced fit model: (the enzyme changes shape to “hug” the substrate)

The enzyme’s shape is not exactly matched to the substrate, but it is able to accommodate the substrate with a close enough shape into an enzyme-substrate complex and carry out catalysing that reaction. Here is a video of an enzyme catalysing a reaction between two molecules into one molecule. This is different to the above scenario in the diagrams, where one molecule is broken down into two molecules.
Enzymes are crucial to proper metabolic function, and ultimately life. That is why you need to know the conditions which affect enzyme activity. There are several properties which can alter enzyme function:
1. Temperature
2. Inhibitors (competitive and non-competitive)
3. pH
4. Substrate concentration.
Enzymes are proteins, so have a delicate tertiary structure that enables that enzyme’s adequate function. High temperature or pH would alter its tertiary structure. Inhibitors would bind to its active site, preventing substrates from doing so. This results in no enzyme-substrate complexes being formed. Let’s have a closer look at these properties individually.
Temperature
Increasing temperature results in a higher rate of activity, up to a certain point where the enzyme becomes denatured. A high temperature causes the molecule to vibrate, breaking the weak bonds that hold it together, and changing the structure of the enzyme. This process is denaturation. The point at which this happens is usually around 50 – 60 degrees Celsius.
Denatured enzymes don’t work. Look at this graph (click to enlarge) to understand the relationship between enzyme activity and temperature:

Inhibitors
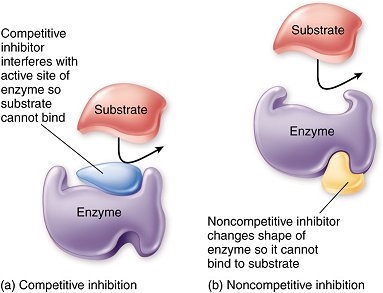
Inhibitors are molecules which interfere with the substrate binding to the active site of an enzyme, slowing down or stopping the reaction. These may be reversible or non-reversible inhibitors. The reversible inhibitors can be competitive or non-competitive.
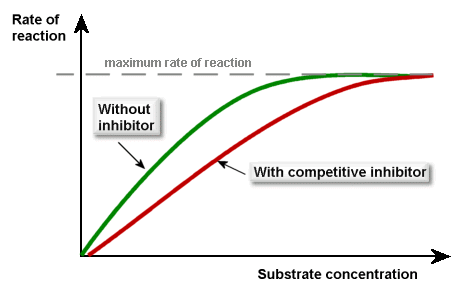
Competitive inhibitors have a similar 3D shape to the substrate, hence they can bind to the active site of the enzyme, preventing the substrate from doing so. It’s easy to picture:

The competitive inhibitors compete (as you’d expect) with the substrate for the active sites of the enzymes. If more substrate is added, then the inhibitors’ effect will be diminished. This is what the graph looks like (make sure you can recall this):
Non-competitive inhibitors on the other hand bind to the enzyme at a site away from the active site. All good? No, because that results in the enzyme’s shape changing. This means the substrate can no longer bind to the active site. Unlike the case of competitive inhibitors, changing the substrate concentration will not have an effect on the rate of reaction. Here is a comparison diagram (learn this):
If you’re a video sort of person, here is a nice one:
pH
Binding of the substrate to the enzyme depends on a close match between shape and charge. The pH is a measure of the concentration of H+ ions versus OH- ions. As you can see, these are positively or negatively charged, so a really high or really low pH can disrupt enzyme function. All enzymes have a specific optimal pH at which they work best. This differs between enzymes. For example, while most enzymes work best at a pH of 7.35 (that is halfway between 1 and 14 – 1 is most acidic, 14 is most basic), pepsin in the stomach acid works best at a pH of 3.
Substrate and enzyme concentration
This topic is a matter of common sense. However, you must use A level language. Here it goes.
Common sense version: More substrate results in more reactions, so rate of reaction goes up. Of course, when all enzymes are working all the time, adding even more substrate will not increase the rate of reaction, unless more enzymes are added.
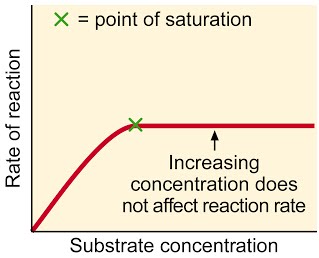
A level language version: The higher the substrate concentration, the faster the rate of reaction until the enzymes are working as fast as possible. This is when all the active sites are filled all the time. From this point, the only way to increase the rate any further is to add more enzyme.
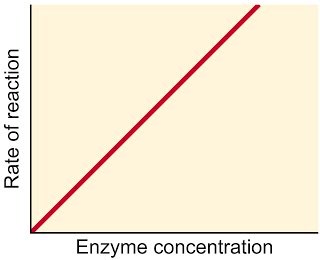
If on the other hand we have excess substrate, adding increasing amounts of enzyme will allow the rate of reaction to increase linearly. What would happen if there were a limited amount of substrate instead? The reaction rate would fall off a cliff as soon as the substrate has been used up.
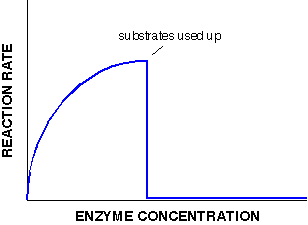
No matter how much enzyme we add, the substrate is no longer present so the reaction rate halts, and no more product can be made. In this case, the initial increase in reaction rate is slower (not linear) compared to the unlimited substrate graph. This is because the substrate is increasingly used up and the reaction rate increases ever more slowly before it stops and drops.
Being able to measure the initial rate of enzyme activity is useful in finding out whether a certain substance acts as an enzyme at all (by observing if adding it to a substrate depletes the substrate or creates a product over time), the efficiency of enzyme activity and hence the affinity of that enzyme to the tested substrate.
For example, we can have two test tubes, one with a lot of starch and one with a lot of lactose (a lot relative to the enzyme we’re about to add; a lot is termed excess substrate). A lot might be 100 or 1,000 starch or lactose molecules for 1 enzyme molecule. We might need to experiment to actually find out how fast the reaction takes place if at all.
We then add an equal amount of enzyme to both starch and lactose tubes and detect how much there is left every minute, or 10 seconds or 5 minutes (again, depending on how fast we expect anything to happen, and that depends on the temperature, etc.).
The results might show that our enzyme doesn’t affect the amount of starch, but completely breaks down the lactose within 12 minutes. The enzyme has high affinity to lactose, hey it might be lactase!
Overall, enzymes catalyse a huge range of both intracellular reactions such as breaking down glycogen to release more glucose for energy, and extracellular reactions such as breaking down environmental components in soil by releasing the enzymes out of the body – this is a form of nutrition in some fungi.
Final tip: you should be aware of the lock and key versus induced fit models. The induced fit model is better because it suggests the enzyme changes shape slightly to accommodate the shape of the substrate. This is beneficial if the enzyme is perhaps fluctuating in shape due to change in temperature for example.
