DNA structure
DNA (deoxyribonucleic acid) is a large molecule which carries the genetic information, or blueprint, of all life on Earth. Mutations arising in the DNA code account for the diversity upon which evolution by natural selection can work. Therefore, it is not far-fetched to say that DNA is one of the central, most important molecules in living organisms.
For such an important molecule, it sure looks beautiful:

DNA is a double helix i.e. two individual strands running along each other in an anti-parallel way, connected to one another by relatively weak hydrogen bonds. DNA’s structure can be learned easily by thinking about the strands and the “stuff in-between” separately.
What are the strands made of?
The strands are made of repeating units consisting of a deoxyribose (sugar) molecule with a phosphate molecule attached to it; hence, it is called a sugar-phosphate backbone.
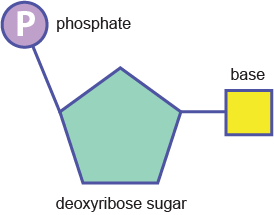
Phosphodiester bonds between nucleotides (above) create the backbone:
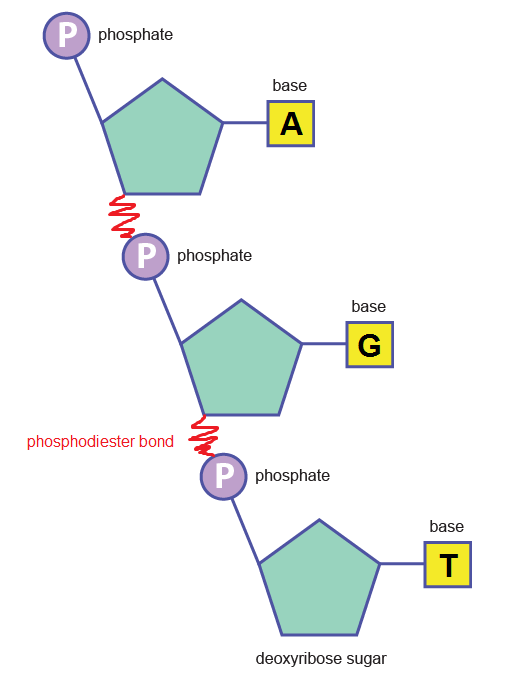
What is the centre made of?
Attached to the sugar molecules in the backbone are a different type of molecule called nitrogenous base. There are 4 bases in DNA: adenine, thymine, cytosine and guanine. These are abbreviated by their initials: A, T, C and G.
The hydrogen bonds are formed between these bases. Due to their complementary shapes, A always pairs with T, and C always pairs with G. A-T is linked by 2 H bonds, while C-G is linked by 3.
Here is a diagram of this arrangement:
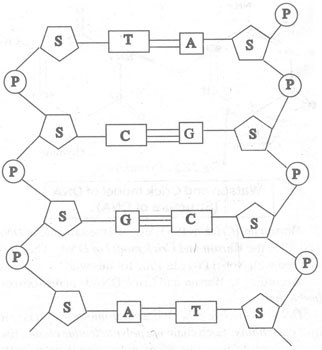
And another:
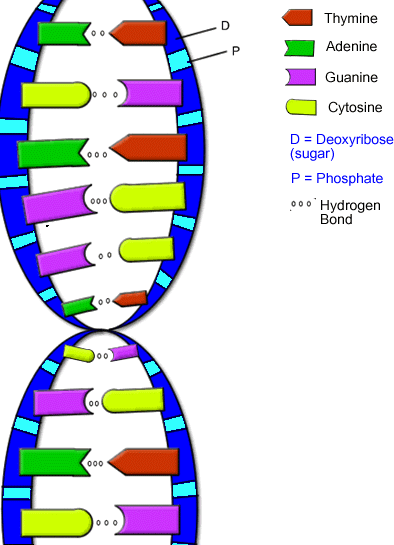
The bases can be sorted into two categories: purines and pyrimidines depending on their ring structure:
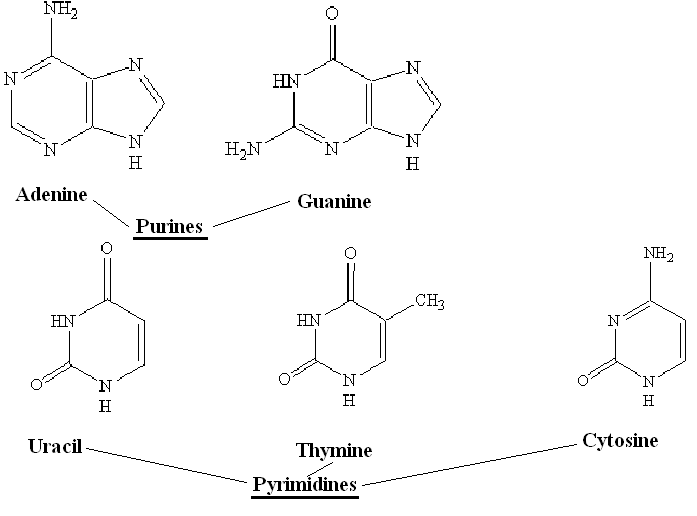
As you can see, adenine and guanine are bigger and have two rings, while thymine and cytosine only have one ring. Uracil is similar to thymine and also pairs with adenine. The presence of uracil instead of thymine occurs in RNA rather than DNA.
DNA is a very stable molecule, as its purpose of carrying genetic information is very important. Features of this are:
1. DNA is very temperature-resistant, and the H bonds only break at temperatures of about 92 degrees Celsius
2. The sugar-phosphate backbone acts as a shield to the bases, preventing interference from outside chemical reactions
3. The double helix gives stability
4. Many H bonds contribute to the stability
5. The structure of the sugar-phosphate backbone itself confers strength.
DNA replication
DNA, of course, replicates. Why? It’s a pretty crucial element in the reproduction of living things. For example, a bacterium replicates by splitting itself into 2 (binary fission). The DNA must stay intact and be copied with a high degree of accuracy in order for the two newly formed bacteria to develop and function as their parent – adequately. In multi-cellular organisms such as ourselves, DNA replication occurs as a prelude to cell division.
For such a complex molecule, past scientists have had a challenging time working out the precise mechanism by which DNA replicates. Three hypotheses were made: (for this purpose imagine one DNA molecule)
1. The DNA molecule replicates by providing itself as a template for a brand new shiny DNA molecule, and then remaining its own intact DNA molecule. This is called the conservative replication model.
2. The DNA molecule replicates by providing itself as a template and being modified itself throughout, resulting in 2 new DNA molecules with patches of the old parent DNA molecule combined with patches of brand new material. This is called the dispersive replication model.
3. The DNA molecule replicates by providing each of its strands as a template for 2 new DNA molecules, each having one entire new strand, and one entire old strand from the parent DNA molecule. This is called the semi-conservative replication model.
Here’s a visual aid for those who found the above descriptions gibberish:
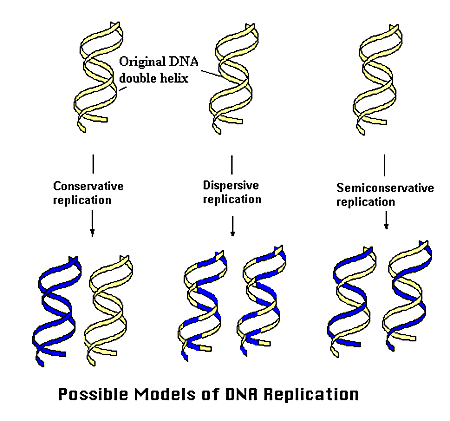
How does one go about working out which one of these models is the correct one? Well, in the ’50s these two chaps by the names of Meselson and Stahl cracked the riddle by carrying out a classic experiment which the examiners are in love with (so learn it well). Such complicated affairs can only be properly depicted by a lovely video. Videos always give the impression that what you are watching, surely, must be a piece of entertainment rather than advanced biology.
Now that we’re all clear on the replication model of DNA, let’s delve deeper into the details of the process. These are the key steps involved in the semi-conservative replication of DNA:
1. The enzyme DNA helicase unwinds the double helix, causing the hydrogen bonds between the two polynucleotide strands to break.
2. DNA-binding proteins maintain the 2 strands separate during replication.
3. Enzymes called primases attach primers to the exposed strand. Primers are a few nucleotides long and constitute the site where DNA polymerase starts its action.
4. DNA polymerase binds to the aforementioned primer and begins catalysing the reaction between free nucleotides (new) and DNA-bound nucleotides (old). Of course this complies with the principle of complemetarity i.e. A-T, C-G.
5. Because one strand is replicated continuously while the complementary strand is replicated backwards, and hence in fragments (Okazaki fragments) rather than continuously, the resulting DNA fragments must be connected together to form the new strand. New phosphodiester bonds between the sugar-phosphate backbone of DNA are catalysed by the enzyme DNA ligase.
The genetic code
Having already covered the basics of DNA, let’s turn our attention to the principles which govern what actually happens to DNA and how this results in life being the way it is!
The Dogma
DNA is a large molecule made up of variable bases (adenine, thymine, cytosine, guanine). The precise sequence and location of these bases determines what structure a second molecule, mRNA (messenger RNA) has once it’s “read” the template DNA. In turn, the sequence and location of mRNA bases determines what amino acids will be chosen in the assembly of a given protein that the original DNA encoded for, once it reaches a ribosome and is constructed by tRNA (transfer RNA).
mRNA
mRNA stands for messenger ribonucleic acid. DNA is deoxyribonucleic acid, and the only difference really is in the sugar in the backbone. A more important difference is that mRNA is single-stranded unlike double-stranded DNA. Additionally, instead of the base thymine, mRNA uses uracil. So while adenine pairs up with thymine in DNA, it pairs up with uracil in mRNA. Knowing that, the mRNA derived from this DNA (looking at the top strand) would be as follows:
DNA: ATGGGTACAAATGC (top strand)
TACCCATGTTTACG (bottom strand)
mRNA: AUGGGUACAAAUGC (single strand)
As you can see, both the top DNA strand and the mRNA are complementary to the bottom DNA strand (in reality either top or bottom may be read, but for simplicity we only look at the top strand whenever it’s given – we assume that is the gene of interest). Therefore the top strand may be called the coding strand (or sense strand) while the bottom is the template strand (or anti-sense strand). It’s called template because it’s the bit of DNA used to actually build up the mRNA according to. The result? The coding strand of DNA except that T is replaced by U!
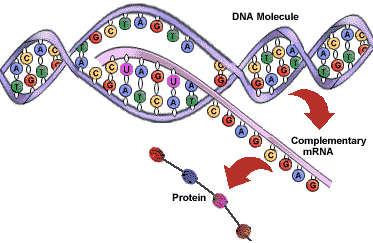
How is mRNA read? An amino acid is coded for by 3 bases in a row. These are called triplets. AUG codes for methionine (Met) which happens to be the amino acid which signals that a new gene starts, if at a certain position within the overall code. Therefore it’s known as a start codon.
The 3 Secrets of mRNA/DNA
There are 3 key properties of the genetic code which regulate its activity.
1. The genetic code is universal. That’s right, the 4 bases are the same in all living things – humans, apples, worms, swans, oak trees, etc.! Moreover, the amino acids coded for by these bases are also completely the same, so AUG codes for the amino acid methionine in all living organisms.
2. The genetic code is non-overlapping, so if you have an mRNA AUGCGA it would be read “AUG”, “CGA” and not “AUG”, “UGC”. The amino acids obtained would be methionine and arginine (Arg).
Tables and diagrams showing you what codes corresponds to what amino acids are widely available and you won’t be expected to memorise them.
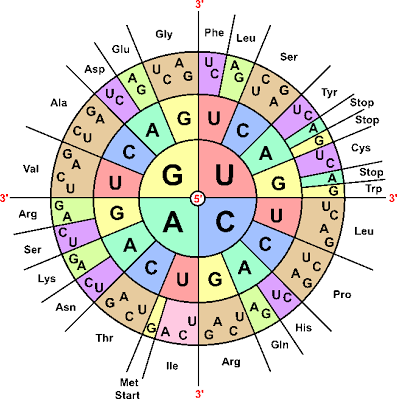
In addition to the start codon methionine, there are multiple stop codons such as UAG and UGA. These signal where the code can stop its translation into the amino acid sequence.
3. The genetic code is degenerate. That might sound slightly offensive, but bear with! Look above, what do the triplet codes UGU and UGC code for (start reading from the inside out by picking each letter)? They both code for cysteine (Cys). How about CUU, CUA, CUC and CUG? They all code for leucine (Leu). This property of different triplet codes coding for the same amino acid is why the genetic code is termed degenerate.
Additionally, some DNA (and in many organisms most of the DNA) does not actually code for amino acids at all. Some repeats many times over, some has regulatory functions, and some has yet to be cracked in terms of its role in the overall function of the organism.
tRNA (Transfer RNA)
We know DNA is double-stranded and uses A, G, C and T bases, while mRNA is single-stranded and uses U instead of T. What about tRNA? Well, tRNA is a very different soup indeed.
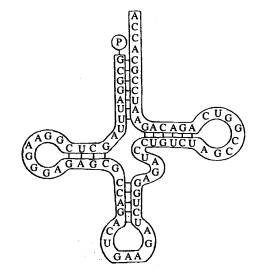
It’s clover-shaped and uses the same bases as mRNA. It is single-stranded, and where one part of the strand meets another there are hydrogen bonds between bases just like in DNA except that in DNA there are 2 strands bonded rather than 2 parts of the same strand).
At the top of tRNA as seen above there is an amino acid binding site (P is seen as attached), while at the bottom there is an anticodon – in this case it’s GAA. The anticodon is complementary to an mRNA codon (triplet code – in this case it would have to be CUU).
Protein synthesis
Proteins are made up of amino acids linked by peptide bonds, therefore a protein may be referred to as a polypeptide (of course, some proteins such as haemoglobin have extra bits to them). All are encoded for by the information stored in DNA. Let’s see how exactly this happens.
Transcription: DNA to mRNA
In a process called transcription, mRNA is formed based on DNA. The bases on the coding strand of DNA are transcribed into a new molecule, mRNA, which is synthesised by the enzyme RNA polymerase.
Wanna see more detail?
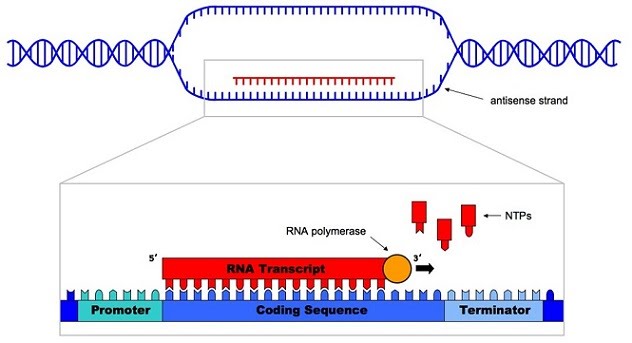
As you can see, the DNA double helix unwinds, RNA polymerase anneals to the coding strand and recruits freely available bases (A, U, C, G) to build an mRNA strand.
Splicing: pre-mRNA to mRNA
In eukaryotes, genes contain non-coding sequences which must be removed before mRNA is used to produce proteins. These are called introns as opposed to exons which are coding sequences. Splicing therefore is the process of excising (cutting out) introns to be left with mRNA containing purely coding sequence.

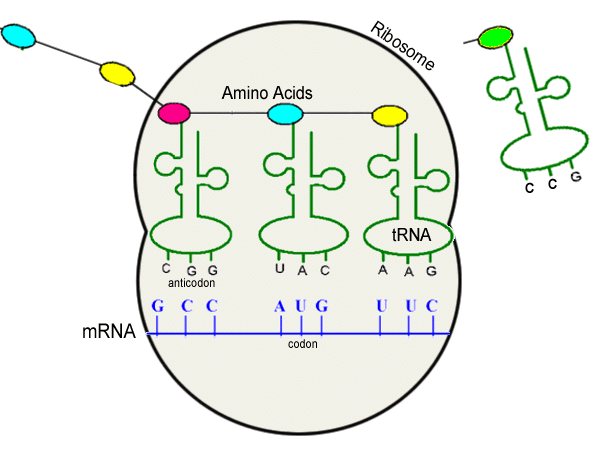
Translation: mRNA to tRNA
The resulting mRNA finally leaves the nucleus where the above business had been taking place, and arrives in the cytoplasm where the final step takes place. More specifically, in ribosomes. Each mRNA codon is matched against an anticodon on tRNA, which is matched to its respective amino acid. This binds to the next amino acid and so forth, until a polypeptide is made.
Mutation
The Cause of Mutations
Mutations are a random occurrence during DNA replication and the rate of mutation is influenced by external factors such as UV radiation. There are different types of mutation:
1. Deletion where a nucleotide base is deleted. AGTCA becomes AGCA.
2. Substitution where a nucleotide base is replaced by another. AGTCA becomes AGTCG.
3. Insertion where a nucleotide base is added as extra. AGTCA becomes ATGTCA.
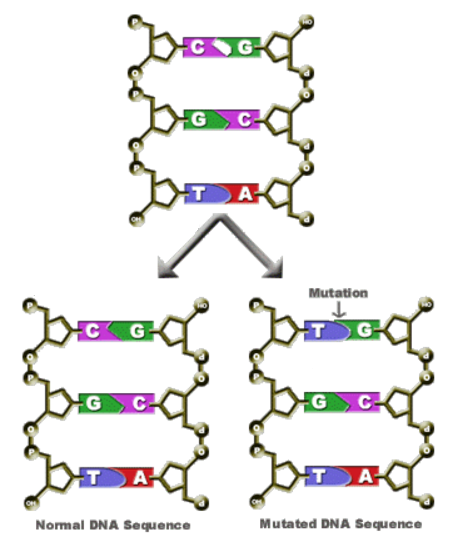
The Effect of Mutations
Since the genetic code is degenerate, it’s possible that a mutation won’t have any effect whatsoever! This represents silent mutations. If 2 different triplet codes translate into the same amino acid, the polypeptide chain will remain unchanged. This of course only applies to substitutions.
Another scenario where a mutation may cause no effect is if it arises in an intron. Since these are removed before mRNA is translated, no mutations would be carried along.
What happens if a base is deleted or added? The genetic code is non-overlapping, so the error cannot simply be overlooked and the following triplets read correctly. The entire subsequent code will be shifted. This is called a frameshift.
Deletion: AGT GGC TTA… –> lose the first G –> ATG GCT TA…
Insertion: AGT GGC TTA… –> insert an A after the first A –> AAG TGG CTT A…
The code is affected significantly!!! In fact, it may be totally ruined. One way this can happen is by a nonsense mutation which by a frameshift causes the code to arrive at a stop codon earlier than it’s supposed to. This will result in a shorter polypeptide and therefore truncated protein which may malfunction.
A missense mutation is when a substitution changes the amino acid encoded. This does not necessarily impact the overall protein, but it may result in a protein with an altered binding site and therefore affect its activity.
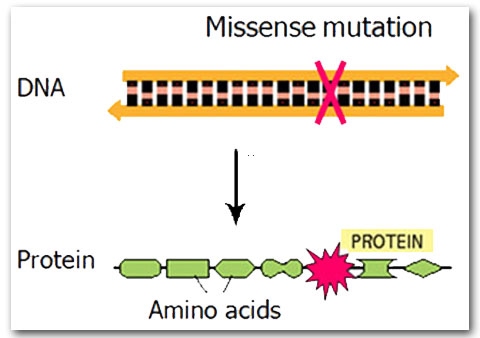
Mutations and Cancer
Cell division is kept in check by two kinds of genes: proto-oncogenes which trigger division, and tumour suppressor genes which inhibit division. Cancer is caused by mutated genes involved in cell division. Mutated proto-oncogenes, called oncogenes, trigger cell division at a far greater rate than normal, thus allowing cells to divide out of control.
Mutated tumour suppressor genes fail to inhibit division any longer, therefore contributing to the growth of cancerous tissue by indefinite division.
Sickle cell anaemia
Sickle cell anaemia is a part of sickle cell disease which is a genetic condition affecting the haemoglobin in our red blood cells. This impairs its function of carrying oxygen in the blood and hence can cause symptoms of anaemia such as dizziness, rapid heart rate and fatigue.
Quite rarely, a condition is caused by a simple point mutation of just one DNA base. This is the case in sickle cell anaemia. The single change, in this case a substitution, happens to result in a different amino acid being coded for altogether, as the codon the mutated base is part of codes for valine instead of glutamic acid in this case.
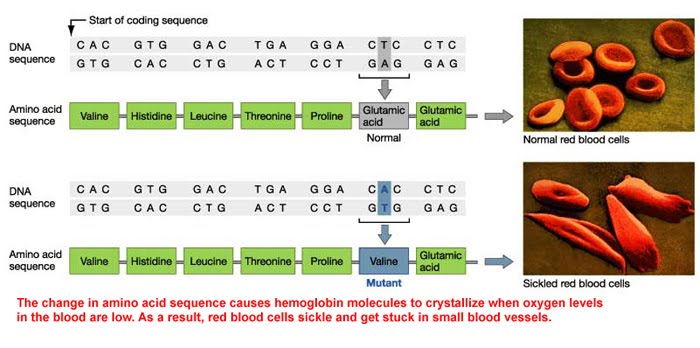
This results in different properties in the new haemoglobin, and as red blood cells contain millions of haemoglobin molecules, it alters the red blood cell structure and function too.
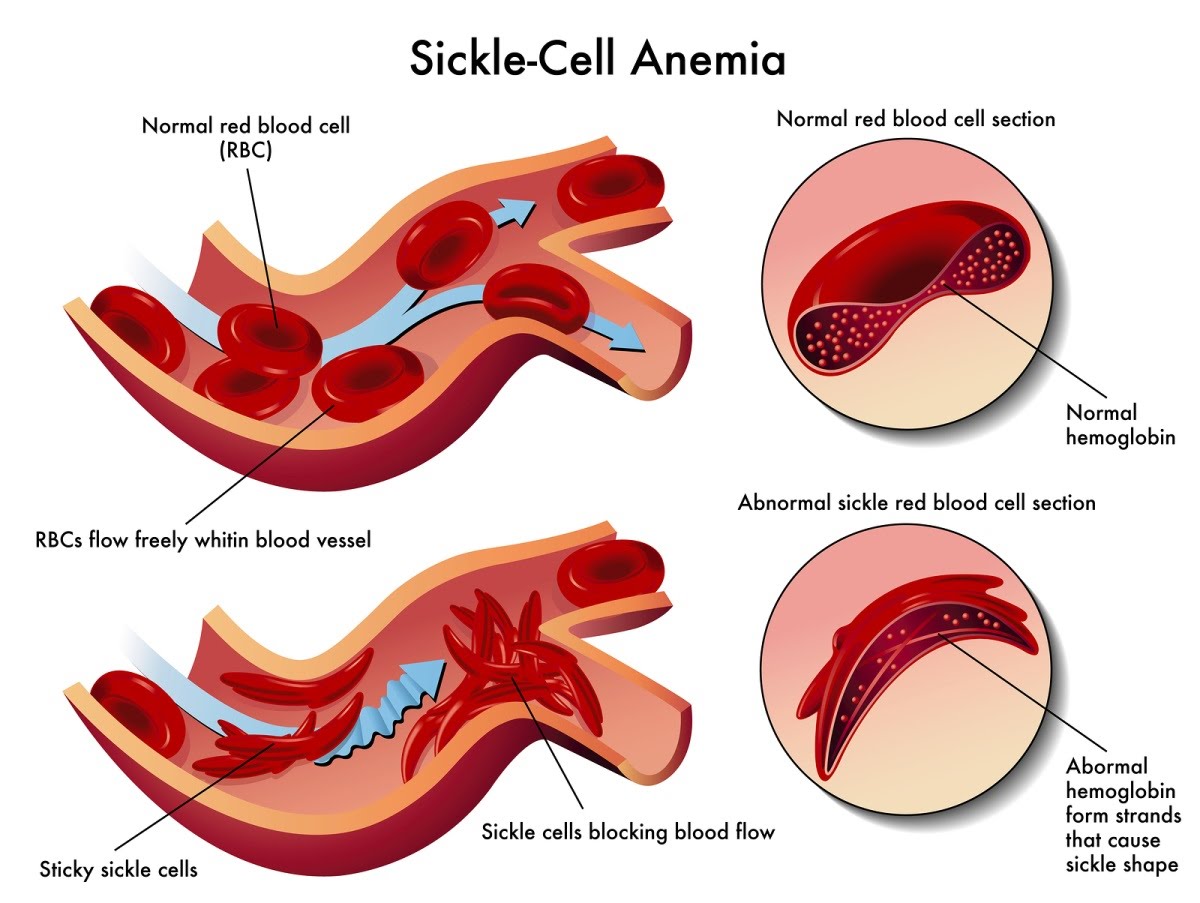
They become sticky and compromise circulation. The different shape of the cell, resembling a sickle, gives this condition its name. In areas where HIV is endemic, sickle cell disease has spread significantly.
Despite it being a detrimental feature, it seems to confer some resistance if HIV infection has taken place in the same individual. In this way the condition has persisted since it is advantageous to those who also have HIV.
Ok byeeeeeeee
